Theme: Modern Exploration in the field of Structural Biology.
Structural Biology Meet 2018
- About Structural Biology Meet 2018
- Sessions & Tracks
- Market Analysis
- Related Conferences
- Past Conference Report
- New Updates: About Structural Biology
On behalf of the Organizing Committee of Structural Biology 2018, all the researchers, developers, and experts are warmly welcome from the field of Structural Biology to attend 10th International Congress on Structural Biology schedule in the month of October 18-19, 2018 in Helsinki, Finland. We aim to unify all the people indulged in this vast field and share the knowledge, explore and look forward to the new way by integrating new thoughts and customizing the limits of the future technology.
Structural Biology 2018 offers a dais to take an attention to explore, grip and meet with prominent speakers of the field, including both broad and specific subjects. The Structural Biology 2018 will be surrounding the theme “Modern Exploration in Structural Biology”.
The Structural Biology 2018 constitutes of Sessions like Keynote Speeches, Oral Presentations, Poster Presentations, Universal Workshops, B2B Meetings, Panel Discussions, Q&A sessions, Industry expert interactions. There are awards for some categories like Best Poster, Best Oral presentation, Young Researcher Forums (YRF), e-Poster presentations, Video presentations by the experts from both Industry & Academic.
International Congress on Structural Biology 2018 invites all interested participants to join us for this venerate event at the elegant destination Helsinki, Finland
WHY TO ATTEND?
International Congress on Structural Biology is amid the World's leading technical Congress. The two days event on Structural Biology will host 60+ Scientific and technical sessions and sub-sessions on leading and latest research transformation in the field of Structural Biology covering the globe. Structural Biology 2018 will comprise of 14 major sessions designed to offer comprehensive sessions that discourse current topics in the field of Structural Biology.
The attendees can find exclusive sessions and panel discussions on latest innovations in Structural Biology by:
-
Lectures from renowned speakers
-
Keynote forums by Prominent Professors, Engineers
-
Open Innovation Challenges
-
Poster presentations by Young Researchers
-
Global Networking sessions with 50+ Countries
-
Novel techniques to benefit your research
-
Best platform for Global business and Networking opportunities
-
Meet the editors of refereed journals, Society and Association members across the Globe
-
Excellent platform to showcase the latest innovation and concept in the technical field
Target Audience:-
-
Biochemists
-
Biotechnologists
-
-
Biochemistry Researchers
-
-
Molecular Diagnosticians
-
Pharmaceutical Leaders
-
Cell Biologists
-
Developmental Biologists
-
Geneticists
-
Micro Biologists
-
Molecular Biologists
-
System Biologists
-
Immunologists
-
Biophysicist
-
Structural biologists
-
Bioengineers
-
Nanoengineers
-
Neuro-Biologist
-
Computational Biologist
-
Biomedical Researchers
-
-
Biomedical and Biotechnology Research organizations
-
-
Structural Biology Societies and Associations
-
Molecular Biology Societies and Associations
-
Professors & Academicians
-
Industrialists
-
Students
Sessions and Tracks
Track 1: 3D Structure Determination
Biomolecules are very small to see in detail even by most microscopes. The methods that the structural biologists use to determine their structures, in general, involve the measurements on huge numbers of identical molecules at the same time. Some of the best methods include X-ray crystallography, Cryo-Electron Microscopy, and Nuclear Magnetic Resonance. Very often scientists use them to study the "native states" of biomolecules. Analytical Techniques are designed for making the qualitative and quantitative calculation. Precision analytical technologies are required to determine product quality and trace level of impurities which may prove to be a risk to human health or the environment. These technologies indulge highly specialized analytical instruments which can only be operated by scientists who have industry application experience.
-
Electrophoresis
-
Gel Chromatography
-
X-Ray Crystallography
-
Electron Microscopy
-
NMR Spectroscopy
-
Immunochemical Techniques For Identification& Estimation Of Macromolecules
Track 2: Computational Approaches in Structural Biology
Computational approaches are benefits for structural biology. Structure of molecules is determined by experimental methods which are tedious and practical. Computational biology is an interdisciplinary field that develops and applies computational methods to analyze large collections of biological data, such as genetic sequences, cell populations or protein samples, to make new predictions or discover new biology. The computational methods used include analytical methods, mathematical modeling, and simulation. It is a rapidly developing multi-disciplinary field. The systematic achievements of data made possible by genomics & proteomics technologies have created a tremendous gap between available data & their biological interpretation.
-
Ab-initio modeling
-
Homology modeling
-
Threading method
Track 3: Hybrid Approaches for Structure Prediction
Structural bioinformatics is especially a practical result for protein structure determination. Structural Bioinformatics is an interdisciplinary field that deals with the three-dimensional structures of biomolecules. It attempts to model and discover the basic principles underlying biological machinery at the molecular level. It is based on the hypothesis that 3D structural information of a biological system is the basic to understanding its mechanism of action and function. Structural bioinformatics combines applications of physical and chemical principles with algorithms from computational science. Major areas protein and nucleic acid 3D structure determination, prediction of protein 3D structure from sequence, protein structure validation protein structure comparison and alignment, protein and nucleic acid structure classification inferring protein function from structure, prediction of protein-ligand interaction, prediction of protein-protein interactions, development of databases.
-
Hybrid of experimental methods
-
NMR structures
-
Hybrid of computational methods
-
Determining protein complex structures
-
Bottom-up integration of atomic detail crystallography
-
Hybrid approaches in complementing high-resolution structural biology
Track 4: Sequencing
Sequencing meets structural biology is a dedicated track to show how the recently developed methods are used to determine the structure of molecules. This approach proves itself helpful in a more efficient way. Synergistic use of three-dimensional structures and deep sequencing is done to realize the effect of the personalized medicine. To know the order of nucleotide in small targeted genomic regions or entire genome sequencing method is utilized. Sequencing allows researchers to ask virtually any question related to the genome, transcriptome, or epigenome of an organism as it enables a wide range of application. Next-generation sequencing (NGS) method is different as for how the DNA or RNA samples are placed, and data analysis is done
-
DNA sequencing
-
RNA sequencing
-
Protein sequencing
-
Polysaccharide sequencing
Track 5: Structural Biology Databases
A biological macromolecule's function is known by the chemical and physical properties of its threeâ€dimensional (3D) structure. For this one should know the structure of a biomolecule which is very helpful if we want to understand the living systems and diseases. A database is a structured collection of data. In the field of structural biology enormous research is being done and as a result, massive data is being produced. In order to pile up the data in an organized manner, bioinformatics databases are used. Many databases are created so that the biological data can be stored such as sequence databases, signaling database, structure database, etc. The Protein database(PDB) which a crystallographic database is used for 3D structural data of larger biomolecules. The advancement in technologies has been reflected in the further development of the PDB and in the structural specialty and structural characteristic databases that have also evolved. In the field of structural biology, the mainly used databases are Protein Data Bank (PDB), Electron Microscopy Data Bank, Protein Structure Classification Database (CATH) and Structural Classification of Protein (SCOP).
-
Classification of structural database
-
Classification of protein structure
-
Protein structure classification database
-
Protein data bank
-
Electron microscopy data bank
Track 6: Signalling Biology
Cell signaling is part of any communication process that governs key activities of cells and coordinates all cell actions. The ability of cells to perceive and correctly respond to their microenvironment is the basis of development, tissue repair, and immunity, as well as normal tissue homeostasis. Errors in signaling interactions and cellular information processing are responsible for diseases such as cancer, autoimmunity, and diabetes. By understanding cell signaling, diseases may be treated more effectively and, theoretically, artificial tissues may be created the process by which a gene's information is converted into the structures and functions of a cell by a process of producing a biologically functional molecule of either protein or RNA is made. Sophisticated programs of gene expression are extensively observed in biology, for example, to trigger developmental pathways, adapt to new food sources, or respond to environmental stimuli. Gene expression can be modulated, from transcription initiation to post-translation modification of a protein.
-
Signaling between cells of one organism and multiple organisms
-
Classification
-
Cell signaling in multicellular organisms
-
Receptors for cell motility and differentiation
-
Signaling pathways
-
Intraspecies and interspecies signaling
Track 7: Molecular Modelling and Dynamics
Molecular dynamics(MD) is a computer simulation method for understanding the physical movements of atoms and molecules. The interaction between atoms and molecules is for a fixed period of time, giving a view of the dynamic evolution of the system. The molecular simulation uses powerful computers to know the interactions between atoms and to understand the properties of materials. Such simulations involve methods that range from very small quantum mechanical calculations on atoms to classical dynamics of large groups of molecules on a timescale of milliseconds or longer. These techniques are used in various fields of drug design, computational chemistry, materials science and computational biology for knowing macromolecular systems ranging from small to large biological systems. Simplest calculations can be achieved manually, but computers are required to perform molecular modeling of the reasonably large sized system. The common feature of molecular modeling methods is the atomistic level description of the molecular systems. This may include treating atoms as the smallest individual unit or explicitly modeling electrons of each atom.
-
Potentials In Molecular Dynamics
-
Steered Molecular Dynamics
-
Molecular Dynamics Algorithms
-
Incorporating Molecular Dynamics
-
Design Constraints
Track 8: Drug Designing
Drug design is an inventive process to find new medication centered on the knowledge of the biological target. It is also known as also known as rational drug design. A drug is most a small molecule that inhibits or activates the function of a biomolecule, which in results in a therapeutic benefit to the patient. Drug design commonly relies on computational techniques. This type of modeling is often mentioned as computer-aided drug design. Drug design that depends on the knowledge of the 3D structure of the target is known as structure-based drug design. The main methods available for drug design are structure-based drug design and ligand-based drug design. The structure-based drug designing also known as direct drug design involves the three-dimensional structure of a drug target interacting with small molecules is used to guide drug discovery. It represents the idea that one can see exactly how the molecules interact with its target protein. Ligand-based drug design also known as indirect drug design is an approach used in the absence of the receptor 3D information and it relies on knowledge of molecules that bind to the biological target of interest. The most important and widely used tools in ligand-based drug design involve QSAR & pharmacophore modeling
-
Rational Drug Discovery
-
Computer-Aided Drug Design
-
Drug Targets
-
Types
Track 9: Frontiers in Structural Biology
The main focus of a structural biologist is protein structure determination and drug design. Protein plays an important role in human body. Proteins are one of the most important parts of any biological systems. Living things would not exist without proteins as it is involved in all aspects of living things. Several proteins provide structure to cells; others tend to bind to and carry vital molecules all through the body. Some proteins are involved in biochemical reactions in the body which are termed as enzymes. Others are involved in muscle contractions and immunity. Structure determination of proteins has always been a challenging field. The complex areas in the field include viruses, pathogens, membrane proteins and signaling pathways. Novel progressions are being done in the arenas of nanopatterning and multiscale modeling of cell signaling proteins. Understanding the folding of the amino-acid chain to produce functional proteins is essential for studying cellular systems. A problem in structural bioinformatics is to determine the three-dimensional (3-D) structure of a protein when only a sequence of amino acid residues is given. These methods can be divided into four main classes: (a) first principle methods without database information; (b) first principle methods with database information; (c) fold recognition and threading methods, and (d) comparative modeling methods and sequence alignment strategies.
-
Primary Structure
-
Secondary Structure
-
Tertiary Structure
-
Quaternary Structure
Track 10: Structural Biology in Cancer Research
The major part of research is being carried out in the area of cancer. The main aim is to design and discover novel and effective drugs to cure the disease. Structural biology combined with molecular modeling mainly aims at drug designing. Eventually, many researchers in Structural biology carry out cancer research to extend the exploitation of molecular understanding of biomolecules in the advancement of novel cancer therapies. Cancer immunotherapy is also used which is defined as the response of immune system to reject cancer. Immunotherapies can be a better way of treating cancer. The malignant tumor cells are attacked by stimulating the immune system as these cells are responsible to exploit the fact that cancer cells often have molecules on their surface that can be detected by the tumor-associated antigens (TAAs).
-
Antibody Therapy
-
Cellular Immunotherapy
-
Cytokine Therapy
-
Combinational Immunotherapy
Track 11: Structural Biology Complexity Arenas
Structural biology focuses at the atomic level for understanding the biomolecules. Most of the aspects of structural biology are complex. Researchers are proving to be successful in solving these complexities like the determination of protein structures, functional annotations and drug designing. Though structures of proteins are solved on a huge scale, the gap between available sequence data and structure data is enormous. Bridging this gap is one of the main challenges. In the current research, some of the most complex areas are protein folding, catching the complication of dynamic nanomachines and signaling networks, understanding the intrinsically disordered proteins.
-
Targeting Intrinsically Disordered Proteins
-
Catching the Complexity of Dynamic Nanomachines
-
Bridging the Gap between Sequence Data and Structure Data
-
Networks of Signaling
-
Protein Folding Dynamics
Track 12: Recent Advances in Structural Biology
Structural biology is one of the developing fields. In the course of time, many innovations have taken place. A large number of solved structures have amplified rapidly. The field of drug design and drug discovery has been advanced. Functional annotations are another field where progressions are being seen. Alterations, in order to improve the effectiveness of prevailing tools, can also be noted. Remarkable advances can be seen in the areas of imaging technologies and advancement of hybrid methods to understand the structure and function of proteins. Structural biology is one of the progressing fields. In the course of time, many developments have been taking place. Huge numbers of solved structures have exaggerated rapidly. The field of drug design and drug discovery has been advanced.
-
Structure Determination
-
Technological Advances In Existing Methods
-
New And Potentially Disruptive Technologies
-
Advances In Drug Design
-
Advances In Tool Development
-
Advances In Imaging Technologies
Track 13: Structural Virology
Viruses use a molecular mechanism to invade the host cells to establish an infection and to ensure that the progeny virus particles are released into the environment invading the host’s immune defenses. This mechanism is known as Structural Virology. Although viruses are as simple as the individual with the self –replicating ability but as a group, they are exceptionally diverse in strategies and structures.
Track 14: Biomolecules
Biomolecules are molecules that are involved in the maintenance and metabolic processes of living organisms. These non-living molecules are the actual foot-soldiers of the battle of sustenance of life. They range from small molecules such as primary and secondary metabolites and hormones to large macromolecules like proteins, nucleic acids, carbohydrates, lipids etc. Biomolecules are usually involved in the maintenance & metabolic processes of living organisms. These molecules are non-living molecules for the battle of sustenance of life. They have a wide range of small molecules like primary & secondary metabolites to Large macromolecules like proteins, lipids, etc. Biomolecules are formed by joining many small units together to form a long chain. This process is called synthesis. These molecules are involved in the maintenance and metabolic processes of living organisms.
-
Types Of Biomolecules
-
Nucleosides & Nucleotides
-
Lipids, Lignin, Amino Acids & Carbohydrates
-
Saccharides
Track 15: Structural Biology and Single Molecules
Single Molecules methods represent a truly novel approach to biochemical/biological problems. All classical structural and biochemistry/biophysics methods describe the behavior of enormous ensembles of molecules, averaging the measured parameters over the entire molecular population. How anyone molecule may behave over time cannot be revealed by such studies; neither can the behavior of individual molecules having different conformations and properties.SM methods provide the only available way to study their functional differences, by recording the behavior of individual members of a certain population of molecules. In addition, SM approaches reveal fluctuations in the observable parameters of a single molecule over time, often with very high temporal resolution, usually on the order of milliseconds.
Track 16: Structural Molecular Biology
Molecular biology is a branch of biochemistry which concerns the molecular basis of biological activity between biomolecules in the various systems of a cell, including the interactions between DNA, RNA, and proteins and their biosynthesis, as well as the regulation of these interactions.
-
Molecular cloning
-
Polymerase chain reaction
-
Gel electrophoresis
-
Macromolecule blotting and probing
-
Microarrays
-
Allele-specific oligonucleotide
Track 17: Structural Bioinformatics and Proteomics
Structural biology determines the 3-D shape of a protein letting us know how a protein functions and the role it plays within a cell. Bioinformatics data is derived from structural determination experiments which provide biological researchers to ask a variety of questions and allowing them to understand how DNA mutations might alter a protein’s shape, disrupt a catalytic site, or alter the binding affinity of a pharmaceutical compound. Proteomics is the large-scale study of proteomes. A proteome is a set of proteins produced in an organism, system, or biological context. The proteome differs from cell to cell and changes over time. The protein activity is also modulated by many factors in addition to the expression level of the relevant gene. Several high-throughput technologies have been developed to investigate proteomes in depth like mass spectrometry-based techniques and gel-based techniques.
Glance at Market of Structural Biology
The concerned region of Structural Biology Analyzer industry market involves North American, Europe, and Asia etc., and the main countries include United States, Germany, Japan, and China etc. North America is the leading region in Structural Biology Analyzers following the European market which is the second largest market for Structural Biology Analyzers. The constant healthcare sector improvements and huge population base represented by the Asia Pacific region is expected to drive the importance in Asia Pacific Biochemistry & Structural Biology Analyzers market.
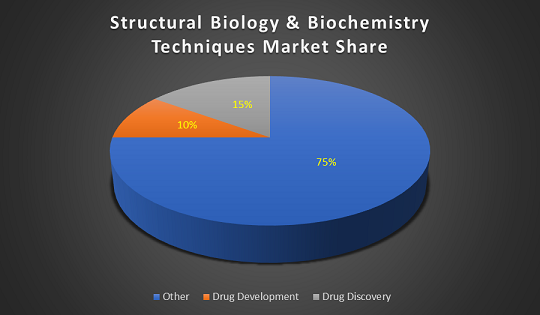
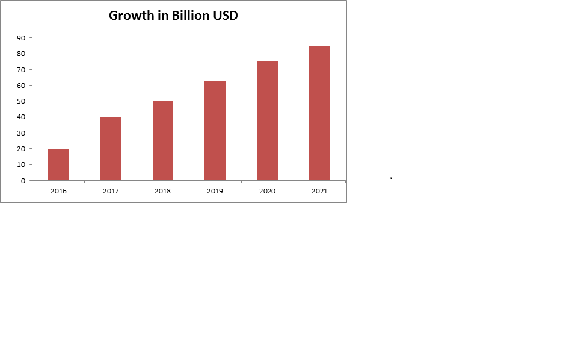
The Structural Biology Analyzers market offers a healthy contribution to the In-Vitro Diagnostic market and is expected to grow in the upcoming years. The global market for in vitro diagnostics (IVD) products was $60.3 billion in 2015 and expected to be $81.1 billion by 2020 at a compound annual growth rate (CAGR) of 6.1%.
North America leads the global IVD products market throughout the period, worth $24.6 billion in 2014. The market is expected to reach $29.4 billion in 2020 from $25.3 billion in 2015 increasing at a CAGR of 3.1%. Asia is the fastest growing region of global IVD market with a CAGR of 12.9% from 2015 to 2020. The market is worth $15.3 billion in 2015 and is expected to reach $28.2 billion by 2020.
Major Structural Biology Related Associations around the Globe:
- American Society for Biochemistry and Molecular Biology
- Czech Society for Structural Biology
- Biophysical Society
- American Chemical Society
- New England Structural Biology Association
- The Protein Society
- American Crystallographic Association
- International Union of Crystallography
- British Crystallographic Association
- European Crystallographic Association
- Hellenic Crystallographic Association
- Bioinformatics society of India
- International Society for Computational Biology
- South African Crystallographic Society
- Australian Society for Biochemistry and Molecular Biology
- Australian Society for Biophysics
- American Society for Mass Spectrometry
- British Biophysical Society
- Bioinformatics Italian Society
- Mid-South Computational Biology and Bioinformatics Society
Major Structural Biology Related Research Units :
- Structural Biology Group, RIKEN
- OKINAWA Institute of Science and Technology
- Markey Center for Structural Biology
- New York Structural Biology Center
- Department of Anatomy and Structural Biology, Einstein College
- Department of Structural and Cellular Biology - Tulane University
- Structural Biology — Penn State University
- Chemical and Structural Biology – The Rockfeller University
- Center for Structural Biology - University of Illinois at Chicago
- Structural Biology Facility - Robert H. Lurie Comprehensive Cancer Center of Northwestern University
- Department of Cellular and Structural Biology – UT Health Science Center
- UCSF Macromolecular Structure Group
- Structural Biology NMR Facility - University of Minnesota
-
12th International Conference on STRUCTURAL AND MOLECULAR BIOLOGY, May 14-15, 2018 Osaka, Japan.
-
10th International Conference on Proteomics, Genomics, and Bioinformatics, May 16-17,2018 Singapore.
-
3rd International Conference on Biochemistry & Molecular Biology, May 16-17, 2018 Singapore City, Singapore.
-
Structural Biology Congress, 2018, 28-30 May 2018, ÅŒsaka-shi, Japan.
-
20th International Conference on Structural Biology, June 21 - 22, 2018, Toronto, Canada.
-
13th International Conference on Structural Biology, July 13-14, 2018, Toronto, Canada.
-
Annual Congress on Advanced Microbiology and Microbiologists, August 01-02, 2018, Toronto, Canada.
-
International Conference on Structural Biology, August 06-07, 2018, Dubai, UAE .
-
World Congress on Structural and Molecular Biology 2018 on August 22-23, 2018 in Rome, Italy,
-
International Conference on Structural Biology and Proteomics, September 3-4, 2018, Bangkok, Thailand
-
International Conference on Computational Biology and Bioinformatics, Sep 05-06 2018, Tokyo, Japan.
-
5th World Congress on Microbial Biotechnology, September 17-18, 2018, Lisbon, Portugal.
-
International Conference on Advanced Microbiology and Research, September 19-20, 2018, Amsterdam, Netherlands.
-
14th International Conference on Structural Biology, September 24-26, 2018, Berlin, Germany.
-
4th International Conference on Biochemistry & Biophysics, October 03-04, 2018, LOS ANGELES, California, USA.
-
13th International Conference on Metabolomics and Systems Biology, October 11-12, 2018, Zurich, Switzerland.
-
13th International Conference on Structural Biology, October 19-20, 2018, Ottawa, Ontario, Canada.
-
12th International Conference on Advancements in Bioinformatics and Drug Discovery, November 26-27, 2018, Dublin, Ireland.
-
11th Edition of International Conference on Structural Biology, March 07-08, 2019, Berlin, Germany.
Structural Biology 2017
The 9th International Conference on Structural Biology was held on September 18-20, 2017 in Zurich, Switzerland with the presence of professional researchers, scientists involved in the development of high-quality education in all aspects.
Structural Biology 2017 witnessed an amalgamation of peerless speakers who enlightened the crowd with their knowledge and confabulated on various new-fangled topics related to the field of structural biology and biophysics. The highly exalted conference hosted by Conference Series was marked with the attendance of renowned and brilliant researchers, business delegates and talented student communities representing more than 20 countries around the world. The conference has tried grounding every aspect related to Structural Biology and Biophysics, covering all the possible research areas and crux.
The conference aimed a parallel rail with the theme “Let's Gear the Future with Structural Biology’’. The meeting engrossed a vicinity of cognizant discussions on novel subjects like 3-D Structure Determination, Computational Approaches in Structural Biology, Hybrid Approaches for Structure Prediction, Structural Biology and Single Molecules, Structural Bioinformatics and Proteomics, Structural Molecular Biology, Structural Virology, Sequencing, Signalling Biology, Molecular Modeling and Dynamics, Drug Designing, Frontiers in Structural Biology, Structural Biology Complexity Arenas and Recent Advances in Structural Biology. The three days event implanted a firm relation of upcoming strategies in the field of Structural Biology with the scientific community. The conceptual and applicable knowledge shared, will also foster organizational collaborations to nurture scientific accelerations.
Cancer Biology
Understanding cancer, and the complex biological systems that underlie its development, is essential if we are to identify new ways of treating the disease. The scientist took a close look at the fundamental mechanisms at work within cells, while also employing complex data analysis and ‘systems biology’ approaches to gain an overview of the intricate webs of communication at play.
The scientists have a proven track record in revealing the biology of cancer. They have made the ground breaking discovery of how the RAS gene, one of the most commonly activated genes in cancer responsible for causing cells to turn malignant.
They identified and understood the BRAF oncogene, which is now an important drug target in malignant melanoma and other cancers.
The science underpinned the development of the PARP inhibitor olaparib, which exploits a weakness in DNA repair in cells with BRCA gene mutations.
Now, guided by the research strategy, the team of scientist continued to lead the way in researching aspects of biology implicated in the origin, growth and spread of cancer.
Scientist having particular expertise in the mechanisms cells use to preserve the integrity of the genome. These systems ensure that healthy cells can only proceed to division if they have first faithfully replicated their DNA.
Their goal was to understand how the mechanisms go wrong in cancer, and ultimately how we can design new treatments that exploit the instability of the genome in cancer cells.
The cancer biologists also have access to the complete range of modern facilities and techniques they need to make discoveries including microscopy, proteomics, metabolomics, advanced structural biology technologies and computational approaches.
And, crucially, fundamental findings by the scientist of cancer biology labs feed into the world-leading programme of drug discovery, helping to pave the way for the next breakthroughs in cancer therapeutics.
A switch to turn fragrances on and off
Researchers have found the switch in plants that turn off the generation of terpenoids which is carbon-rich compounds playing part in plant physiology and are utilized by people in everything from fragrances and flavorings to biofuels and pharmaceuticals.
Plant terpenoids are found in wholesome supplements, natural insecticides, and drugs utilized to treat malaria and cancer. The chemotherapy sedate Taxol, which is utilized to treat breast, ovarian, lung, bladder and prostate cancers, may be a plant terpenoid. But plants frequently make them in such low amounts that extracting them for such uses is expensive and regularly impractical.
A new enzyme was discovered which is available in all plants called isopentenyl phosphate kinase (IPK) that directs the recede and flow of living, carbon-based molecules called terpenoids. Firstly, the role of this enzyme was unraveled in completely different organisms, bacteria and a very ancient group of life called Archaea. By elucidating the three-dimensional structure and chemistry of the enzyme, it was revealed that a previously unknown gene found in all plants encoded the same enzyme as originally discovered in microbes.
As terpenoids utilizes considerable amounts of carbon and energy in plants, it had been recognized that their formation must be under tight control so that they are produced only when important for the bacterium or plant hosts. To unravel how plants, switch on and off metabolic pathways by controlling the ebb and flow of terpenoid production for regulating the availability of their chemical starting materials, scientist in various field came together.
The team had earlier determined how plants turn on terpenoid production but understanding both the "on" and "off" the yin and yang switches--as well as the bottlenecks for flux are essential for understanding and ultimately tuning up terpenoid yield.
This is important basic knowledge that opens new targets for engineering of terpenoid metabolic pathways. Plants usually produce these compounds, but the amounts produced is very small. It might have taken many of the plants to get enough of a compound to use it for something like a pharmaceutical. This new set of unanticipated discoveries will lead to faster, more efficient ways to obtain enough amounts of these products for the benefit of humans.
Scientist have converted chemical pools of inert monophosphate terpenoid building blocks into readily used diphosphate building blocks. Using a multi-pronged approach that incorporates structural biology, biochemistry, plant genetics and synthetic biology, the research group determined that two Nudix enzymes were the missing links responsible for the evacuation of a phosphate group to return the dynamic terpenoid diphosphates back to the dormant pool of terpenoid monophosphates.
The Nudix hydrolase family of enzymes are preserved in all living beings, however, their biological parts are unclear. But researchers have uncovered an unexpected and new function for members of this family in plants.
They have shown that IPK and Nudix are working together to regulate downstream terpenoid product formation. Some of these products may be toxic to the plants if the plants make too much of them. This is how the plants regulates their output.
A technique to identify charge distribution on biomolecules
Researchers have identified a technique to detect, measure and analyze biomolecules with complicated charge distributions. Clinicians often measure biomolecules, with focus frequently being on antibodies. This is because the small proteins attach to antigens; foreign substances that induce immune responses in the body. As most biomolecules have inhomogeneous charge distributions, and as such erratic responses are occasionally received from the traditionally used carbon nanotube systems.
The researchers demonstrated the method, which adjusts the solution that the biomolecule is monitored in and uses carbon nanotube thin film transistors (CNT-TFTs) to precisely identify the exact amount of a specific biomolecule in a specimen. Aptamers are immune antibody receptors used by the CNT-TFTs. They detect the net electric charge of the target biomolecule.
After a biomolecule is identified by the scientists, an antibody is made to attach to it in solution. The antibody then connects to the aptamer on the thin film of carbon nanotubes that converts the connection into an electric signal. This signal is then able to be detected by a sensor.
The scientists are then able to identify the Debye length, determining the distance between a point charge and the molecule therefore mapping a molecules uneven charge distribution. The researchers recognized that they had to look at how the charges were distributed at the surface level of the molecule to understand the behavior in the sensor signal. They stated that despite being the same target molecule, the polarities of the sensor response are different from positive or negative. The improvement of dynamic range was achieved by using a low concentration of buffer solution.
Several different characteristics in an experiment have been shown to influence a molecules Debye length, and the researchers suggest that these results show promise for further controlling sensors and modifying their dynamic range.
The researchers are assuming to apply the findings in real-life scenarios and generate a sensing technology that can be detected under high concentration conditions close to blood.
Advances in biomolecular mass spectrometry (MS) have had a significant impact on the field of structural biology
Understanding the intricate structures of the proteins in our bodies is key to advancing precision medicine. To do this, it’s necessary to look beyond individual proteins and delve into the assembly and structure of protein complexes.
Progress in biomolecular mass spectrometry (MS) has had a significant impact on the field of structural biology. Technology progressions in mass analyzers are the driving constraint behind the expanding number of structural biology investigations, which are permitted by the increased speed, sensitivity, selectivity, and variety of MS fracture methods. This, in turn, has led to a plethora of MS methods, particularly at the intact protein and peptide levels, which allow the characterization of biomolecular structures.
At the intact protein level, native MS allows the study of protein assemblies in their original state by examining the noncovalent protein-protein and protein-ligand complexes. At the peptide level, liquid chromatography (LC)-MS/MS analysis of proteolytic digests provides the amino acid sequence of proteins, allowing protein subunits to be identified from a proteome database. Limited proteolysis and surface labeling techniques such as hydrogen-deuterium exchange MS (HDX-MS) have been employed to monitor conformational changes and characterize protein-protein interfaces. A combination of chemical linking of amino acid residues within a native complex with MS analysis of cross-linked peptides (XL-MS) can determine topological arrangements and reveal where the protein domains interface.
Recently, cryo-electron microscopy (cryo-EM) has emerged as an alternative to traditional techniques such as X-ray crystallography and nuclear magnetic resonance (NMR) imaging. CryoEM can directly visualize complete macromolecular complexes instead of just selected parts. As with MS advance, recent progressions in cryo-EM test planning, microscope and detector technology, information collection automation, and image processing have made it plausible to reproducibly reach near-atomic levels of resolution. Defining the structure of these huge dynamic complexes needs integrating different complementary methods, such as MS and cryo-EM density maps—an approach known as integrative structural biology.
One such example uses structural proteomics MS tools to examine the stoichiometry of KaiA, KaiB, and KaiC which are the components of the cyanobacterial circadian clock and to monitor the well-defined assemblies, followed by structural characterization using single-particle cryo-EM.
The further applications of these synergistic approaches, integrative structural biology has a bright future in accelerating the knowledge and understanding of even more intricate systems such as pathways and organelles along the path from structure to function.
Watching DNA being read with cryo-EM
Researchers have shown the pictures of the DNA code which was readable and interpretable for depicting the new detail about one of the basic forms of life. They used an advanced form of Cryo-electron microscopy to zoom in the image and capture the reading mechanism in unrivalled detail.
They freezed and then catched the RNA polymerase III complex, separating and reading the DNA code. They obtained almost a million of independent snapshots and using powerful computers grouped together the similar snapshots, enhancing the detail to obtain a detailed reconstruction of this machinery at work.
Now, it is known how the components of the RNA polymerase III molecular mechanism fit together and thus, allowing the scientist to design drugs that turn the system on or off which offers a whole new way of treating cancer.
RNA polymerase III is necessary for life of all eukaryotic cells and in cancer, it is more dynamically causing cells to deliver bigger numbers of the building blocks that must be develop and duplicate. Using cryo-EM, the pictures of small molecules i.e approximately 5nm or 20000th of the width of a human hair were taken easily. Cryo-EM involves freezing and imaging samples at -180°C to preserve minute details of protein structure. Cryo-EM is transforming molecular and cellular biology that allow us to investigate minute detail at molecular machines within the cells and their working mechanism. The technique is helpful for scientists in discovering the weaknesses in cancer cells to target them by the next generation of drugs.
What is Bioinformatics?
Bioinformatics – an application of the informatics techniques to obtain, store, and interpret large quantities of biological data. The term Bioinformatics is the study of informatic processes in biotic systems. Its applications are found in different fields of biology among which few are enlisted below:
1.Genomic Sequencing
The study of genomics involves the analysis of genomes, identification of their structure, function, and evolution. In 1990, the Human Genome Project was started with the aim of fully sequencing the human genome via Sanger sequencing. This method created many fragments of DNA that had to be combined using biotechnology processes to create a final complete sequence. With further development in sequencing technologies, an entire genome can be sequenced in a short span of time through a method termed “next-generation sequencing”. This method is an extremely high-throughput, systematic DNA, and cost-effective sequencing technology allowing a huge volume of DNA strands to be sequenced at a time as compared to the Sanger method of genome sequencing that uses fragment-cloning methods.
2.Cancer Research
Another major arena that benefits from bioinformatics is cancer research. Cancer bioinformatics is emerging as an important strategy in systems clinical medicine to improve the patient outcome. Cancer bioinformatics is playing a vital role in the identification and validation of network to dynamic network biomarkers. Network biomarkers comprise the protein-protein interactions. Any changes in these network biomarkers can be monitored at the different time interval of disease progression which is termed as dynamic network biomarkers. The dynamic network biomarkers are linked to the clinical information of a patient such as imaging, biochemical profiling, disease and therapy history of the patient, and other measurements. The biomarkers can help in monitoring the disease progression in a patient. Depending on the disease progression, the existing therapy can either be changed or continued to improve the quality of life of the patient.
3.Personalized Medication
By identifying individual mutations involved in a disease, personalized medicines can be developed. The genetic information obtained from genome sequencing helps in identifying the precise gene where a mutation has taken place and based on which personalized treatments can be developed. The primary relapse analysis demonstrates tumor progression and the case of secondary malignancy which indicates the probability and importance of real-time molecular profiling in the patients to improve individual prognosis.
4.Evolutionary Biology
The study of evolution involves knowing the origins of the species, identification of their ancestors and the possibility of any mutations during the evolutionary process. The first genomic sequence- Phage Ф-X174 was discovered in 1977. Bioinformatics helps in the comparison of numerous species by their genetic profile and helps in the identification of evolutionary trees known as phylogenetic trees. The huge amount of data produced is well organized that allows previously undetected genetic events to be characterized now such as identification of horizontal gene transfer.
Bioinformatics is also used in the analysis of proteins. The structure and function of proteins are characterized using various high-throughput technologies that allow the scientists to gain a greater insight into their roles in the biological processes. The field of bioinformatics has revolutionized the analysis of data in many areas of biology allowing us to seek answers for the most important and fundamental biological questions without the burden of accumulating a large amount of data.
The Effect of Protein Structure on Disease
Proteins are the biochemical compounds which consist of polypeptides which are folded into a globular or fibrous form in a biologically functional way. They form the body’s most essential building blocks. They control all of the body’s functions and are referred to as the cell’s molecular engines. Their actions are diverse and complex and depend on their precise three-dimensional (3D) structure.
The structures of proteins allow seeing the biological processes at their most fundamental level, through knowledge of the 3D structure, which helps in identifying which proteins, interact and how does it happen, the mechanism of drugs, and how certain diseases proceed at an atomic level.
The recent advancement allows the 3D structures to be used to design ‘smart drugs’. This approach utilizes the knowledge of the structure of a protein to customize the design of drugs to interact with it, thus having the potential to shave years and millions of dollars of the traditional drug-design approaches.
The studies also contribute to the identification of targets for cancer therapy along with the understanding of the mechanisms of cancer growth. Mainly, the focus is on a protein complex called the GM-CSF/IL-3/IL-5 receptor, a cell signaling receptor in the blood control system. Malfunctions in the signaling pathway result in diseases such as certain types of leukemia. Their discovery helps to explain how this receptor is activated and will form the springboard for the development of new treatments.
Much of the knowledge of disease comes from an improved understanding of the functioning of proteins hence giving the knowledge about the best possibility of designing new drugs and treatment strategies.
To gain insight into the underlying mechanisms of various biological events, it is important to study the structure and function correlation of proteins within cells. Thus, to diagnose the diseases related with the biomolecules like protein, structural biology plays a major role in determining the protein structures.
New clue to Huntington’s disease
Insulin signaling could slow the disease’s progression
A study reveals that it is achievable to restrict the advancement of Huntington’s disease on increasing insulin signaling within the brain’s neuronal cells. The experimentation conducted was a follow up to investigations that had uncovered that the insulin signaling pathway which is a critical controller of cellular metabolism and energy homeostasis was severely compromised in Huntington’s and different polyglutamine disorders like SCA3.
On increasing the level of insulin signaling in Drosophila, the cellular pools of proteins which are essential are enriched for cellular functioning and survival. The cellular transcription machinery which collapses due to the disease can also be restored and thus preventing the disease from progressing.
Still without a cure
Huntington’s disease is still not curable. The infected individual’s losses the ability to walk, talk, think and reason. The disease initiates between the ages of 30 and 45 and each individual with the gene for the disease will eventually develop the disease. It is an autosomal dominant genetic disorder i.e. if one of the parents carries the defective Huntington’s gene; their offspring has 50% chances of inheriting the disease.
On initiating the experiments to know if the increased level of insulin signaling would increase by regulating the insulin receptor in the disease affected neuronal cells. Hence it was found to be useful. The researchers then tried to reveal the molecular mechanism and found that there is a significant reduction in the cellular level of neurotoxic protein aggregates or inclusion bodies, with a remarkable decrease in the neuronal cell death.
The enhanced level of insulin signaling restores the neuronal cells which are otherwise stressed during disease condition. The study says that anti-diabetic drugs can be efficiently used to restrict the pathogenesis of polyglutamine disorders.
Therefore, the study has immense potential application and its subsequent investigations by including higher organisms and anti-diabetic drugs would provide a novel approach to combat the devastating human polyglutamine disorders.
Machine learning classifies biomolecules
Recently a machine learning-based method to classify biomolecules using existing Small angle X-ray scattering (SAXS) data has been published. Small angle X-ray scattering (SAXS) is one of a number of biophysical techniques used for determining the structural characteristics of biomolecules. This method is used to classify shape as well as to estimate structural parameters like the maximal diameter or molecular mass of the molecule under study. These estimates may then serve as a valuable method for validating expected values.
Some parameters are decided for the shape classifications for biomolecules: flat discs, compact-hollow cylinders, extended rods, compact spheres, hollow spheres and flat rings. They used simulations to obtain idealized scattering profiles of each of these different geometries across a range of heights, widths, and lengths ranging from 10 to 500 Å.
- Shape classifications
Simulated scattering intensity profiles for different molecular shape classes:
(a) The reduced form of the intensity profile for each shape class
(b) Each shape class defines a point cloud in normalized apparent volume space, with each shape falling into well-defined regions
The innovative data reduction approaches are used to reduce each of the scattering profiles to a point in normalized apparent volume space denoted by V. It was advantageous to represent the data in this way as the structures sharing similar structural characteristics will occupy a similar position in V space.
The unknown scattering profile for classification then amounts to calculating its position in V space and locating the nearest points in V space for which parameters are already known. The new parameters can then be estimated by taking a weighted average of these “nearest neighbor” points in V space. A machine can be programmed to perform all of these steps.
- Using machine learning
The huge amount of scattering patterns about 488,000 was simulated to train an algorithm to categorize different scattering patterns. Each scattering pattern was then removed and the remaining data were used to predict the shape classification of the removed pattern.
This training procedure allowed the refining of the weights assigned to the nearest neighboring structures in V space, so as to maximize the accuracy of the machine classification.
- Predicting structural parameters
To test the predictive power of the shape classification method, the scattering data from the Protein Data Bank (PDB) and the Small Angle Scattering Biological Data Bank (SASBDB) was harvested.
From the atomic structures stored in the PDB, they used CRYSOL software to generate scattering intensities, as well as values of structural parameters such as the maximal diameter and molecular mass. After mapping the known structures to V space, an equivalent algorithm was then used to predict the structural parameters based on the generated scattering intensity. Here, the machine prediction was within 10% of the expected value in 90% of cases.
The SASBDB provides scattering intensity as well as user-generated values of structural parameters such as the maximal diameter. It was also observed the good agreement from the structures collected from the SASBDB, with the machine predicting a small, systematically lower value for the maximal diameter. This offset reflects the fact that molecules tend to occupy an extended configuration in solution.
The protocol developed shows that data mining has significant potential to increase the efficiency and reliability of scattering data, which could have the huge benefit for the biophysics community.
Pandoravirus: Giant viruses invent their own genes
Pandora-viruses appears to be as big as bacteria and containing genomes that are more complex than those found in some eukaryotic organisms i.e. organisms whose cells contain nuclei, unlike the two other kingdoms of living organisms, bacteria and archaea. Their strange shape and vast typical genome (up to 2.7 million base pairs.) led scientists to wonder where they came from.
The scientist's team has isolated three new members of the family compared those six known cases using different approaches. On being analyzed it showed that despite having very similar shapes and functions, the viruses only share half of their genes coding for proteins. However, members of the same family have more genes in common.
The new members of virus family contain a large number of genes referred as orphan genes which encode proteins that are not equivalent in other living organisms as in the case for the two previously discovered Pandora-viruses creating a new reason for debate.
Bioinformatics analysis reveals that the orphan genes exhibit features which are similar to those of non-coding regions in the Pandora-virus genome. This is the only possible explanation for the gigantic size of Pandora-virus genomes, for their diversity and the large proportion of orphan genes as they contain most of the virus’s genes that might originate spontaneously and randomly in non-coding regions. Usually, genes appear at different locations from one strain to another, thus explaining their unique nature.
On confirming this hypothesis it will make these giant viruses of genetic creativity which is a central, but still rarely explained the component of any understanding of the source of life and its evolution.
The bridge between epigenetic and cancer treatment
Not every person who has a "cancer gene" will inevitably develop this disease in their lifetime. This is due to a new area of research called epigenetics, where it is known that many genes are only read under certain circumstances. Scientists have not only researched the underlying mechanisms but also have developed small molecules that inhibit the derailment of gene expression in cancer.
Many seminal contributions have been made to the elucidation of structural mechanisms of the regulation of proteins that play key roles in signal transduction. This, in turn, led to specific regulatory mechanisms, a detailed description of protein family and the elucidation of substrate recognition processes. Based on these findings, a large array of new small molecule inhibitors, are developed in particular highly specific inhibitors for epigenetic reader domains.
The primary developed potent inhibitor approves epigenetic pursuer spaces as a focus for malignancy treatment which prompted an assorted arrangement of profoundly particular inhibitors focusing on the proteins. This work has given new vision into chromatin science and made the way for more than 25 clinical trials in this new region of drug discovery.
Proximity labeling—dissecting gene transcription's proteomic choreography
A new approach which is more powerful than previous methods for tagging and harvesting DNA-associated proteins from cells could open deeper insights into transcription control.
Gene transcription requires a complicated feat of molecular choreography, in which numerous proteins come together in the right place at the right time. Isolating, identifying, and studying those proteins and existing methods for doing so is similarly complicated while powerful have limitations that restrict their utility.
The widely-used chromatin immunoprecipitation (ChIP) approach relies on antibodies to fish proteins out of a cell. It can only find proteins for which one has a high-quality, specific antibody and that one assumes may be present near a given gene, leaving out proteins that are unexpected.
A new unbiased approach for capturing proteins that help express a gene of interest, one that relies on a blunted version of the CRISPR-Cas9 genome editing system. Dubbed GLoPro (for genomic locus proteomics), the approach provides a more comprehensive view of which proteins manage a given gene's expression, the knowledge that can help reveal insights into its function and its place in the cell's circuitry.
GLoPro relies on a catalytically inert (or "dead") form of the Cas9 protein (dCas9) fused to an engineered version of an enzyme called APEX2. Using guide RNAs, the fused protein—called CASPEX—homes in on a desired site in the genome in typical CRISPR fashion. Instead of cutting DNA, however, CASPEX labels, on command, any proteins or groups of proteins in its proximity with biotin. Researchers can then use this chemical tag to collect the labeled proteins for mass spectrometry-based proteomic analysis.
In cell lines, researchers could label, capture, and analyze proteins bound to the promoters for the genes hTERT and MYC. Benchmarked against other protein isolation methods, GLoPro proved itself an effective and potentially generalizable way for collaring proteins near any location in the genome. The scientist continues to explore the ways in which GLoPro could serve as a tool for gaining more comprehensive insights into the coordination of gene transcription.
New compound helps activate cancer-fighting T cells
Scientific studies have identified the new mechanism which is responsible for improving our immune system’s activity as it offers new approaches for cancer treatment & vaccination. Chemists have developed a new lipid antigen; stimulating disease-fighting T-cells of the immune system as they create a new path for development of better cancer therapy drugs & vaccines.
According to the studies, the invariant natural killer T-cells(iNKT) acts as a weapon on which our immune system relies to fight against infection and combat diseases like sclerosis, cancer, etc. There are several compounds that show stimulation of iNKT cells in mice but this activity is limited towards human iNKT.
The new compound named AH10-7 had shown similar properties that had been searched by researchers. This compound is the modified version of synthesized ligand which is effective in activating the human iNKT cells. It is selective as it encourages iNKT cells to release a specific set of proteins i.e Th1 cytokines which are responsible for stimulating the anti-tumor immunity. This study had involved advanced structural and 3-D computer modeling.
A crystallized form of the molecular complex was exposed to high-intensity X-ray beam due to which a detailed 3-D image was obtained showing the interaction between killer T-cell receptor and AH10-7. The natural and synthetic form of glycolipid ligands known as alpha-galactosylceramides were potent activators of iNKT cells which showed a positive response in fighting cancer and other diseases. These ligands serve as tiny dockmasters in our immune system, helping antigen-presenting cells attract and bind with iNKT cells so they can be activated to kill cancerous cells or fight off pathogens and other foreign invaders.
Two significant modifications to ligand were made. It was found that adding a hydrocinnamoyl ester on to the sugar stabilized the ligand was kept close to the surface of the antigen-presenting cell, thereby enhancing its ability to dock with and stimulate human iNKT cells.
Molecular details of protein crystal nucleation uncovered
Protein crystal nucleation is a process with great medical & scientific relevance. The protein crystal had been essential for structural biologists to predict the 3-D structure of protein besides this they are also used as bio-pharmaceutical delivery agents. Due to their long shelf life, low viscosity & slow dissolution rate, the crystal suspensions show attraction to store & administer the active pharmaceutical compounds
Despite their tremendous potential, there are two factors that limit the use of protein crystals in a broad range of applications.
First, growing protein crystals for many proteins, crystallization can be very difficult. This is the reason why scientists don't understand the early stages of protein crystal formation. The crystal originates from a nucleus which is a tiny crystalline form to form a spontaneous grouping of molecules in solution to form 3-D structure.
Secondly, a single protein can crystallize in multiple different crystal forms i.e polymorphism due to which different crystal polymorphs show different characteristics. Till yet it is very difficult to know the crystallization process to the polymorph of one's liking.
The scientist had used cryo-transmission electron microscopy (Cryo-TEM) to capture the origin of a protein crystal by visualizing the process of nucleation at molecular resolution. Then the images obtained for the sample at a specific time interval were observed to know the molecular collision that leads to crystalline nucleus formation. Later, it was observed that the protein had a hierarchical self-assembly process. It was then compared with the nucleation pathways of multiple polymorphs and analyzed the difference in structure which was achieved by gently tweaking the different modes of interaction that exist between the molecules, steering the nucleation process
This new insights and methodology will significantly advance the development of protein crystals for 3D structure determination and medical applications.
Artificial cellular signaling models
For cells to perceive its environment & respond accordingly Signaling systems are required so that homeostasis & proper development occurs. Errors in these systems result in many diseases including cancer & diabetes emerging the need for many powerful tools to know the study for development of therapies.
Traditionally, to study cellular signaling pathways biochemical & molecular biology methods were used focusing on individual pathway components. With the advancement in technologies like genomics & proteomics, biology has facilitated a more comprehensive view for signaling.
To know about the molecular machines involved in signaling cascades, a detailed view of their architecture and interactions is required. Often, the low endogenous abundance or tissue heterogeneity of certain proteins obstruct their extraction, purification and structure prediction for which Scientists often make choices to recombinant production and purification in heterologous host cell systems.
Mostly medicines marketed by the pharmaceutical industry are targetting signaling cascades to treat diseases. The EU-funded SynSignal consortium observed that novel tools are needed to study signaling systems relying on synthetics biology techniques to overcome certain product development challenges. Synthetic cellular signaling circuits are recognized as being analogous to electronic circuits. They designed an individual signal building block, assembling them in-vitro to produce synthetic cascades that resemble the natural process.
A DNA sequence of defined structure and function encodes the individual component of the circuit which is physically interchangeable with compatible modular building blocks. It was focused that activation of G-protein-coupled receptors initiates signaling. These pathways also served as a screening platform for new medicines to treat diseases.
Cryo-electron microscopy reveals the structure of a herpesvirus capsid
The reconstruction of the 3.1 Å structure of the herpes simplex virus type 2 (HSV-2) B-capsid & building the atomic model has helped researchers to understand the assembly mechanism of the capsid. This virus is the most complex virus genetically as well as structurally & hence it spread within the host population efficiently causing a wide range of diseases in humans.
In the assembly pathway of these viruses; three distinct types of capsid are produced namely A-, B-, C- capsid respectively having mature angular shells with similar assembly mechanism. Merging “block-based” reconstruction with accurate Ewald sphere corrections helped to reconstruct 3.1 Å structure of the herpes simplex virus type 2 (HSV-2) B-capsid. Among the four layers, the capsid of the virus not only protects the genome from damage but also functions to release it into the host cell nucleus during an early stage of infection & during maturation for packaging of a genome.
It was found that there are four major conformers of capsid protein VP5 exhibiting the difference in configuration and mode of the assembly to form intermolecular networks. Triplex is a heterotrimeric assembly cementing the capsid together; consisting two copies of VP23 & one copy of VP19C. VP26 is a small capsid protein & its 6 copies form a ring-like structure on the top of the hexon; stabilizing the capsid.
Based on the capsid structure, a model was proposed for assembly of the capsid utilizing a triplex leading to the twofold symmetry confirmation on clustering the basic assembly units into a higher-order structure. The nascent assembly intermediates are guided into the correct T = 16 geometry, leading the first step towards understanding the drivers of assembly & the basis of the stability of the capsid.
The radical ways sunlight builds bigger molecules in the atmosphere
With the approach of summer, "sea and sun" might conjure up images of a beach trip. But for scientists, the interactions of the two have big implications for the climate and for the formation of tiny droplets, or aerosols that lead to clouds. The molecules at the ocean surface activate other molecules as an effect of sunlight which results in larger molecules and may affect the atmosphere.
These organic molecules start reacting as they get activated on absorbing sunlight and undergo through reactive intermediate known as “radical” which initiates a chain reaction forming more complex chemicals. The pathway of this "radical initiator" is important to understand which molecules at the sea surface end up in the atmosphere. The molecules which are found in the atmosphere on aerosols determine whether they will absorb or reflect sunlight thus affecting the temperature of the earth. Till now much of the focus was on the hydroxyl radical which efficiently reacts in the atmosphere. It is proposed that a class of compounds; α-keto acids gets photo-activated by sunlight and drives reactions with molecules that do not absorb sunlight themselves.
The two different α-keto acids were studied which showed that light caused the acid to react with several fatty acids and alcohols. Mostly these molecules are found near the ocean's surface and are ubiquitous in biology. It was explained that this sunlight-initiated chemistry has the capability to change the composition of the sea surface. The new and larger molecules formed may add to aerosols hence changing their properties and leading to interesting and previously unforeseen consequences to human health, visibility and climate.
Bioinformatics bridges computer and life sciences
Bioinformatics is a technology which is used to analyze the larger amount of data like human genetic codes. It is a combinational field including chemistry, biochemistry, biology, statistics, and mathematics. To understand biological data, new methods and software tools are also created by Bioinformatics professionals. This project mainly focuses on functional genomics investigating that which pieces of the genome make people who they are.
Another project named Gene Weaver is conducted which has been on-going for the past seven to eight years. It establishes an online toolkit for people to upload and analyze sets of genes, like those related to diseases or biological processes, to find the common genetic components across species.
The program was at first intended to give learning in different fields including informatics, computational science, life science, quality and genome item sequencing and structure examination. Life sciences such as genetics and immunology are also incorporated under Bioinformatics. Informatics incorporates database design and web interfaces, while computational sciences focus on algorithms and modeling.
The several algorithms as part of the research make the predictions about the proteins because they are difficult to predict and classify. A new research project utilizes artificial intelligence algorithms and machine learning to classify proteins and to find their structure and characteristics. Machine learning will be used in future to know cancer with research by finding the patterns of the genome that causes cancer or autoimmune diseases.
Artificial intelligence is kind of new electricity in the industry without it nothing will be completely done in the future.
Scientists create nanomaterial’s that reconfigure in response to biochemical signals
Biological cells have the complex and supernatural ability to recompose and change the way they communicate with each other over time, allowing them to lightly direct critical functions in the human body. A major challenge in materials science is developing nanomaterial that can replicate aspects of these cellular functions and integrate with living systems. Researchers detail how they have created synthetic materials with the ability to mimic some behavior normally associated with living matter.
The ability to self-assemble, rearrange and disassemble in response to chemical signals is a common trait in biological materials, but not in manmade oneself. If we require to integrate synthetic materials into biology, a seamless interface is desirable, which requires materials that share some of the properties of living matter. This approach opens the door to manmade materials that can interact with and repair living systems.
To develop nanomaterial that rearranges in response to chemical signals, researchers started with the base molecule naphthalenediimide (NDI), which is an organic semiconductor. The molecule was selectively modified on both sides by exposing it to biochemical signals in the form of simple amino acids that were added to the system. An enzyme was used to incorporate the amino acids onto the core molecule, triggering self-assembly and disassembly pathways. This process allowed the formation and degradation of nanomaterial with wire-like features capable of conducting electrical signals.
Researchers were able to direct the development of nanomaterial with different properties by using different amino acids, including a programmable nanostructure with the ability to turn electrical conduction on and off through the use of time-dependent self-assembly and disassembly.
These materials exhibit a remarkable ability to remodel their electrical connections just like neurons in the brain. The assembly of these molecules is encoded in their dynamic chemistry; therefore by simply changing the chemical inputs, we are able to observe insulating nanomaterial, conductive nanomaterial, or nanomaterial that dynamically switches between conducting and non-conducting states. The fact that their assembly and conductivity evolve in water makes these materials all the more compelling for bio-interfacing applications.
The devices were developed for measuring the nanomaterial's electrical conducting abilities. The next step is to interface the new nanomaterial with actual neurons to see how the manmade and biological materials interact.
There is still an exciting breakthrough to demonstrate the possibility for creating manmade materials that mimic a complex, dynamic activity of biological systems. This new nanomaterial has the ability to respond to biologically relevant chemical signals and provide an electronic interface. In the long run, this may open up a new pathway towards developing treatments that, until now, have only been theoretical.
Effect of DNA inserts length on whole-exome
Exome sequencing has revolutionized clinical research and diagnostics. Whole-exome sequencing (WES) currently allows the identification of the genetic basis of disease for 25%–40% of patients. A key element of WES is high-quality library preparation and target enrichment.
Typical exome sequencing pathways involve library construction from purified DNA enriched with the exon regions following sequencing. Targeted enrichment can be useful in a number of situations where particular portions of a whole genome need to be analyzed. As sequencing and sample preparation technologies develop, the cost of exome sequencing has reduced substantially. However, the preparation of libraries for target enrichment and sequencing is still complex and sensitive.
Several techniques for optimization of library preparation can be proposed. For example, improve assemblies, save money, accurate size selection can boost sequencing efficiency, and even allow sequencing of low-input samples. In turn, optimal insert size is determined by limitations of the next-generation sequencing (NGS) instrumentation and by specific sequencing application. DNA extraction was performed using QIAamp DNA Mini Kit according to the manufacturer’s instructions. The quality of genomic DNA was verified using electrophoresis on agarose gel. At this stage, lack of DNA degradation and RNA contamination were monitored. DNA concentration was measured using a Qubit 3.0 device. DNA libraries were prepared. Double barcoding was performed by polymerase chain reaction (PCR). The quality control of obtained DNA libraries was carried out using Bioanalyzer 2100. To target the enrichment of the coding regions, the target enrichment system SureSelect XT2 was used. DNA was sequenced on Illumina MiSeq and HiSeq 2500 using pair-end 100 bp reads.
The results indicate that 250–330 bp DNA fragments demonstrate the highest enrichment efficiency. For exome sequencing, about 80% of human exomes on each chromosome are <200 bp in length which is the optimal length for whole-genome sequencing. Therefore, the determination of size selection is an important step for effective enrichment and subsequent sequencing
Conference Highlights
- 3D Structure Determination
- Computational Approaches in Structural Biology
- Sequencing
- Hybrid Approaches for Structure Prediction
- Structural Biology Complexity Arenas
- Structural Biology Databases
- Molecular Modeling & Dynamics
- Drug Designing
- Structural Biology in Cancer Research
- Structural Virology
- Frontiers in Structural Biology
- Recent Advances in Structural Biology
- Biomolecules
- Signalling Biology
- Structural Biology and Single Molecules
- Structural Molecular Biology
- Structural Bioinformatics and Proteomics
To share your views and research, please click here to register for the Conference.
To Collaborate Scientific Professionals around the World
| Conference Date | October 18-19, 2018 | ||
| Sponsors & Exhibitors |
|
||
| Speaker Opportunity Closed | Day 1 | Day 2 | |
| Poster Opportunity Closed | Click Here to View | ||
Useful Links
Special Issues
All accepted abstracts will be published in respective Our International Journals.
Abstracts will be provided with Digital Object Identifier by









































































